muon is a Python framework designed to work with multimodal omics data. Incentified by recent advances in acquisition of multimodal data from individual cells, muon aims to provide convenience and speed to its users enabling standardised analysis while staying flexible and expandable. muon stands on the shoulders of and integrates with annotated data object specification and scanpy library for single-cell analysis in Python.
Application highlight
Analysis of single‑cell RNA & ATAC data from peripheral blood mononuclear cellsApplication highlight
Analysis of single‑cell RNA & surface protein measurements from peripheral blood mononuclear cellsPotential application
Analysis of trimodal single‑cell data – RNA, ATAC, epitopes – from peripheral blood mononuclear cells
get to know muon
multimodal data containers
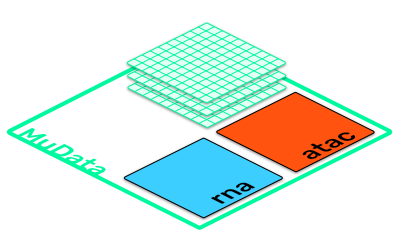
muon introduces multimodal data containers allowing Python users to work with increasigly complex datasets efficiently and to build new workflows and computational tools around it
~> python
mdata
| MuData object with n_obs × n_vars = 10110 × 110101
| 2 modalities
| atac: 10110 x 100001
| rna: 10110 x 10100
multi-omics methods
muon brings multi-omics methods availability to a whole new level: state-of-the-art methods for multi-omics data integration are just a function call away
# multi-omics factor analysis
mu.tl.mofa(mdata)
# weighted nearest neighbours
mu.pp.neighbors(mdata)
methods crafted for omics
muon features methods for specific omics such as ATAC-seq and CITE-seq making it an extendable solution and allowing for prospective growth in an open-source environment
# TF-IDF transformation
ac.pp.tfidf(mdata.mod['atac'])
# denoising and scaling by background
pt.pp.dsb(mdata.mod['prot'])
Get muon
❯
pip install muon
# or
❯
pip install git+https://github.com/scverse/muon
Get started
~> python
import muon as mu
Load data easily whether you start from raw count matrices, Loom files or AnnData objects. Or Snap files. Or Seurat objects. Or MultiAssayExperiment objects. And make multimodal containers with it.
~> python
mdata = MuData({
'rna': adata_rna,
'prot': adata_prot
})
Generate an interpretable latent space leveraging information from multiple modalities. Or cluster your data based on multiple layers of information. Or construct a neighbourhood graph on multiple modalities.
~> python
mu.tl.mofa(mdata)
Visualize results in and across modalities.
~> python
mu.pl.mofa(mdata, color='CD45RA')
mu.pl.umap(mdata, color='celltype')
get involved
From biomedical observations to novel single-cell assays. From chromatin accessibility to spatial transcriptomics. Applications of multimodal data are far-reaching, and it's a community effort.